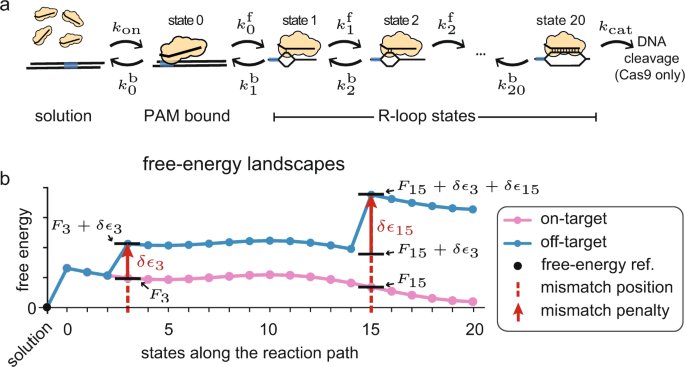
A kinetic model predicts SpCas9 activity, improves off-target classification, and reveals the physical basis of targeting fidelity

Massively parallel evaluation and computational prediction of the activities and specificities of 17 small Cas9s
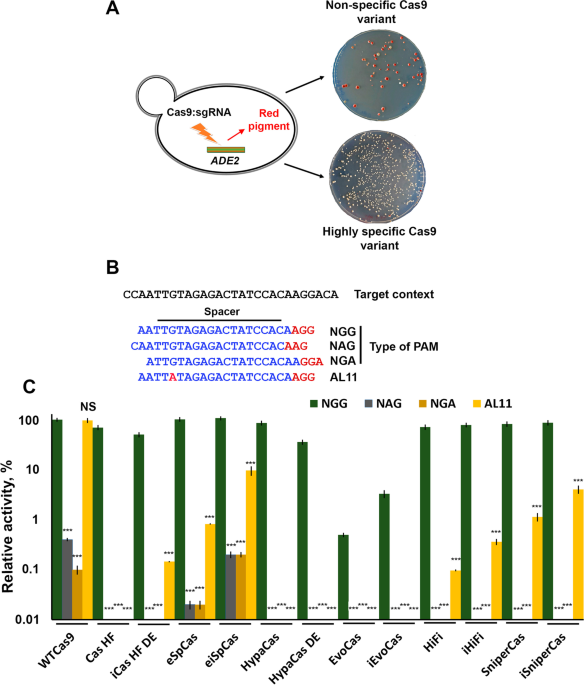
Improving the on-target activity of high-fidelity Cas9 editors by combining rational design and random mutagenesis
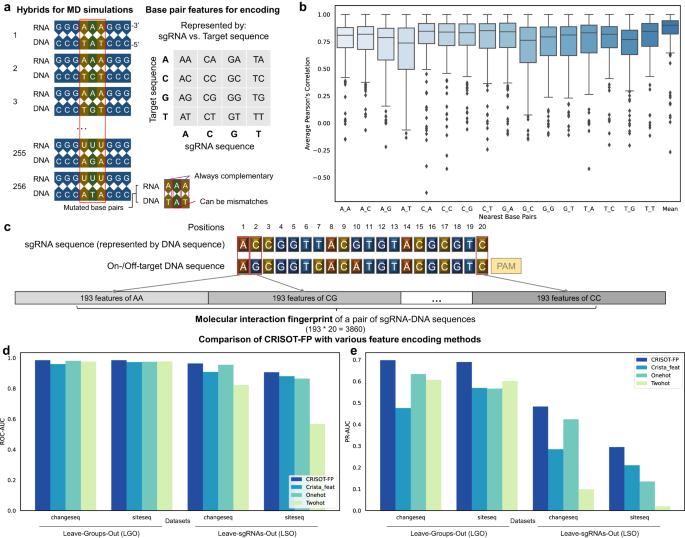
Genome-wide CRISPR off-target prediction and optimization using RNA-DNA interaction fingerprints

Improving the on-target activity of high-fidelity Cas9 editors by combining rational design and random mutagenesis

Next-generation forward genetic screens: uniting high-throughput perturbations with single-cell analysis: Trends in Genetics

PDF) A kinetic model predicts SpCas9 activity, improves off-target classification, and reveals the physical basis of targeting fidelity

A quantitative model for the dynamics of target recognition and off-target rejection by the CRISPR-Cas Cascade complex

PDF) Genome-wide CRISPR off-target prediction and optimization using RNA-DNA interaction fingerprints

A quantitative model for the dynamics of target recognition and off-target rejection by the CRISPR-Cas Cascade complex

Altered DNA repair pathway engagement by engineered CRISPR-Cas9 nucleases

Discovery of Diverse CRISPR-Cas Systems and Expansion of the Genome Engineering Toolbox

Insights into the Mechanism of CRISPR/Cas9-Based Genome Editing from Molecular Dynamics Simulations

Engineered Staphylococcus auricularis Cas9 with high‐fidelity - Wei - 2023 - The FASEB Journal - Wiley Online Library









